|
I am student at UC Berkeley studying in the college of Electrical Engineering and Computer Sciences (EECS) and Molecular-Cellular Biology (MCB). At Berkeley, I am currently doing research in the Berkeley Artificial Intelligence Research (BAIR) Lab, working with Amir Bar, Ph.D. and Professor Trevor Darrell. I am also in the Mobile Sensing Lab, working with Alben Bagabaldo, Ph.D. and Professor Alexandre Bayen. In addition, I am a part-time research intern at the Newman Lab, collaborating with Jeremy D'Silva, Ph.D. and Professor Aaron Newman. Previously, I was an undergraduate researcher at the Sandler Neuroscience Building at UCSF, where I was advised by Dr. Roland Henry. |

|
|
I am fascinated by the advanced computational techniques, with a specific focus on generative modeling and self-supervised representation learning. This passion drives my pursuit of innovative approaches to analyzing and interpreting complex datasets, aiming to uncover novel insights and applications. |
|
A couple of my past research work that are (or soon to be) publicly published and available to see: |

|
I co-founded EgoPet, an extensive dataset comprising over 84 hours of egocentric videos activities and behaviors of various animals. This project involved tasks such as object detection and frame assessment, crucial for improving locomotive performance in the Unitree A1 robotic dog and mirroring animal intelligence and interactions. Our findings and methodologies culminated in a publication submitted to the IEEE / CVF Computer Vision and Pattern Recognition Conference (CVPR). |
|
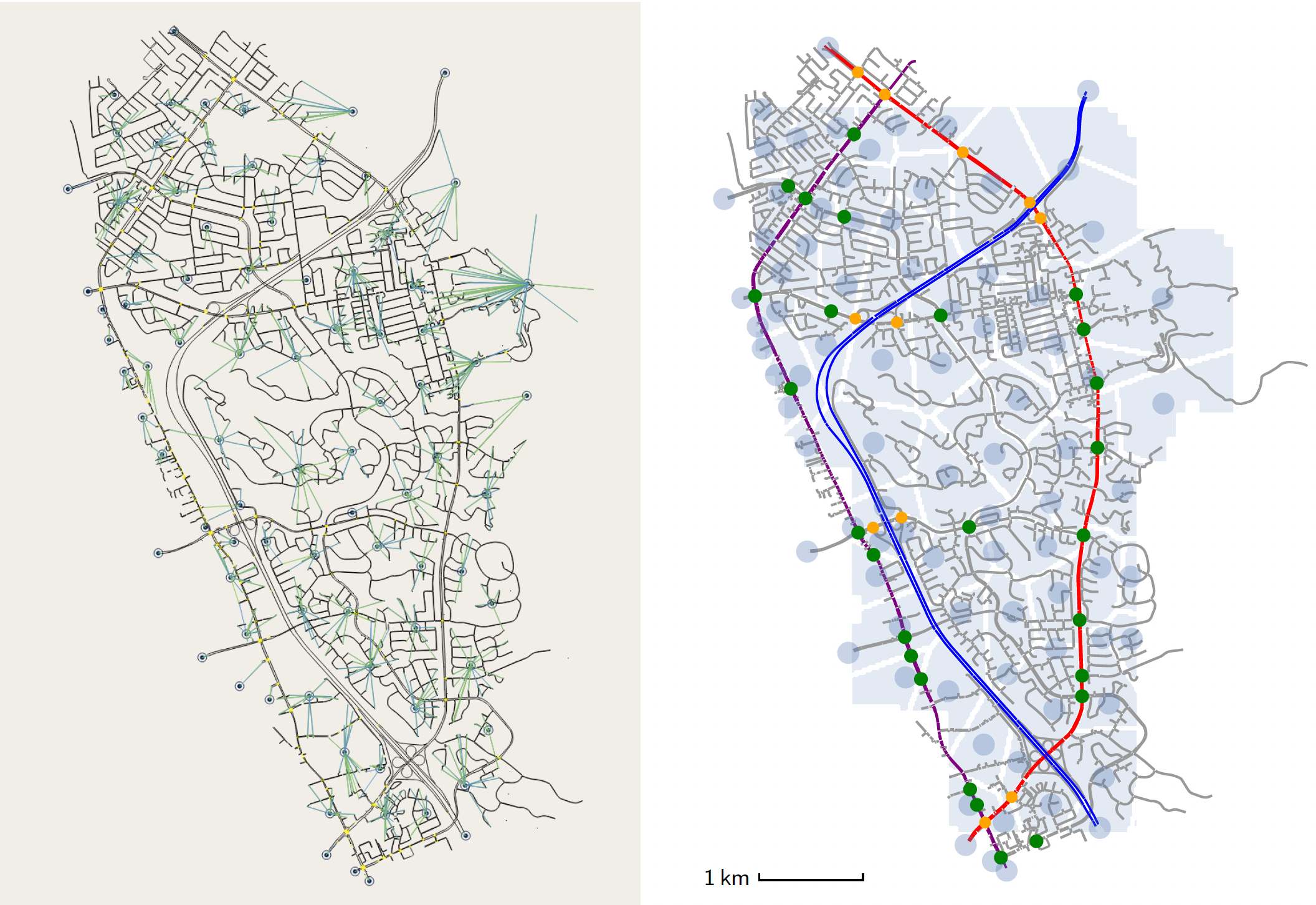
I investigated traffic signal timings in the Mission San Jose District of Fremont, California, using a validated and calibrated microscopic traffic simulation model considering 2019 demand data. Employing a dynamic traffic assignment microsimulation model with a logit-based route choice model, I examined the effects of both uncoordinated signal timings and the strategic reconfiguration of traffic signals on Mission Boulevard, one of Fremont's primary arterial roads. Through my analysis, I discovered strategic reconfigurations of certain signal coordination strategies that could significantly redirect traffic flow, effectively alleviating congestion at targeted intersections and enhance the overall network performance. |
|
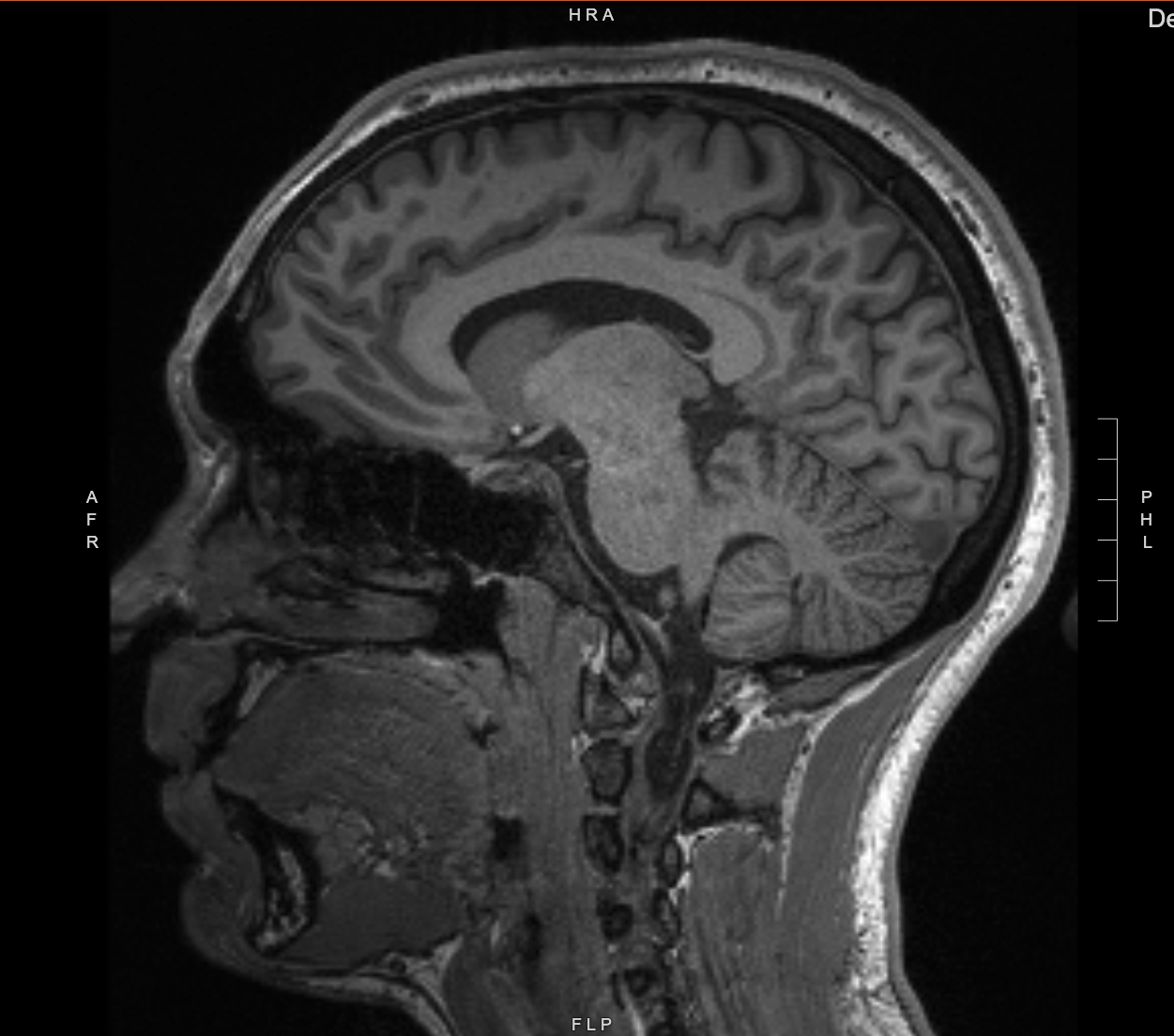
I won a summer SURF fellowship by submitting a proposal to pursue an independent project. I developed software to track the concentration of specific metabolites, such as glutathione, using MRI images collected from patients diagnosed with Multiple Sclerosis. I presented part of my research findings to a summer research cohort. |
|
Some projects that I am continuing to work on: |
|
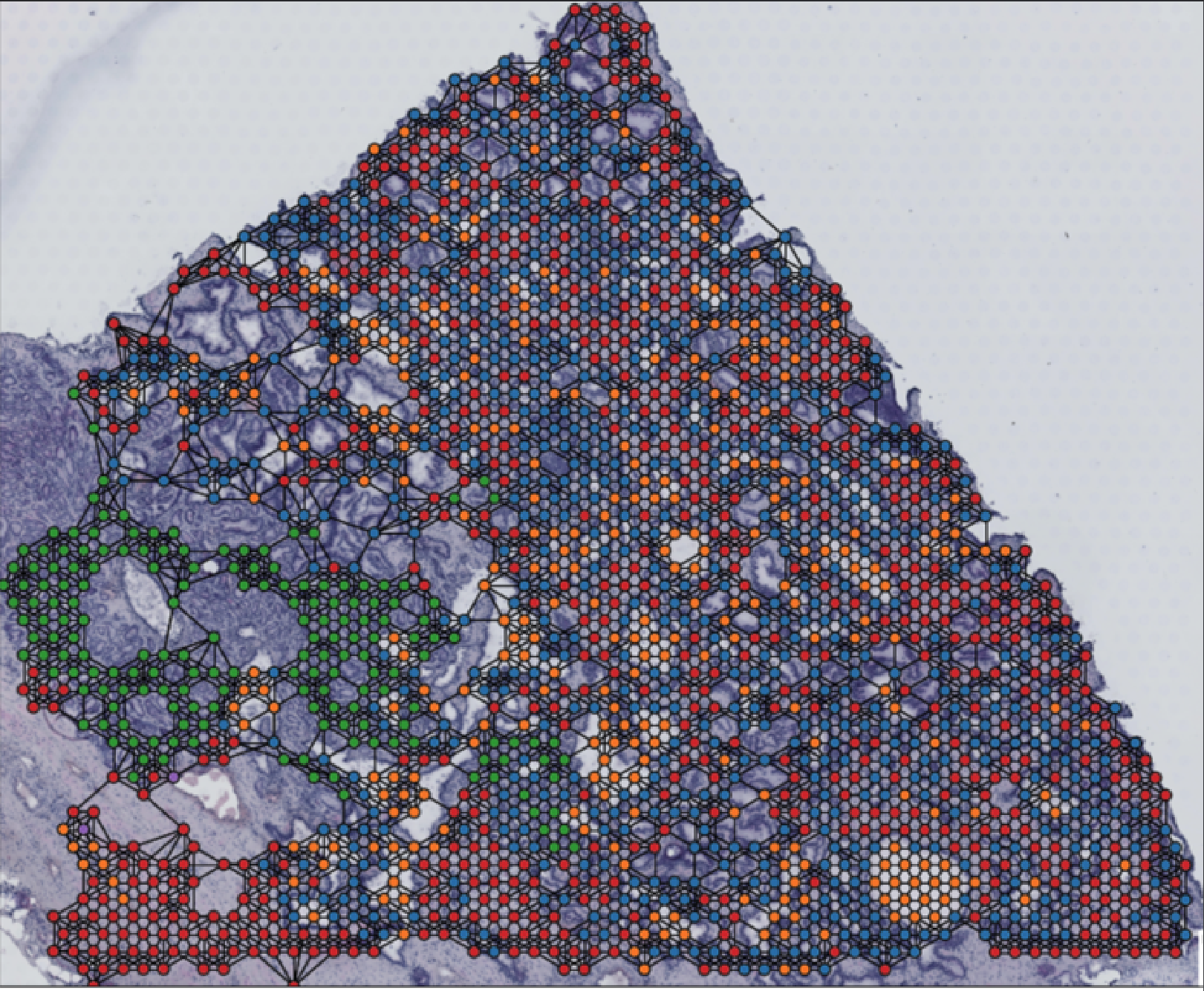
I am investigating the spatial organization of distinct cell types in the tumor microenvironment and its impact on cell state and tumor progression. Utilizing Graph Neural Networks (GNNs), I am modeling complex cell interactions within tumors, capturing nuances in cellular behavior. My work primarily employs PyTorch Geometric, a tool for deep learning on graph-structured data, to analyze and interpret these cellular networks. Through this approach, I aim to unravel the intricate dynamics of tumor cells and their environment, contributing to advancements in cancer treatment strategies. |
|
I am conducting data extraction and enhancement on single-cell RNA sequencing (scRNA-Seq) information sourced from academic publications that studied diverse patient groups. Utilizing both R and Python, I am developing scripts to perform comprehensive analyses on how individual cell differences influence patient characteristics. In addition, I am working in close partnership with two doctoral candidates, taking an active role in periodic updates for the GlyScope initiative. |
|
|
|
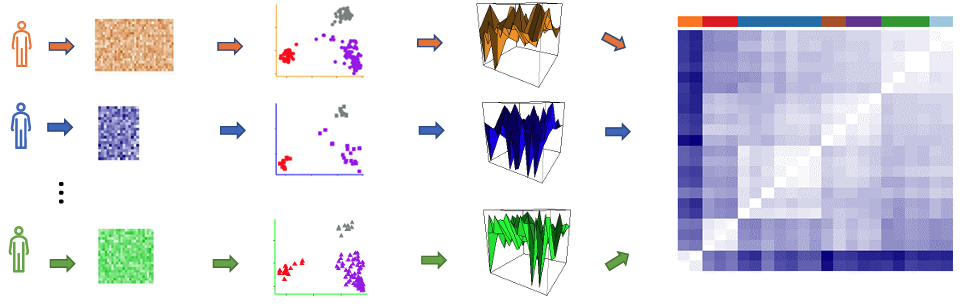
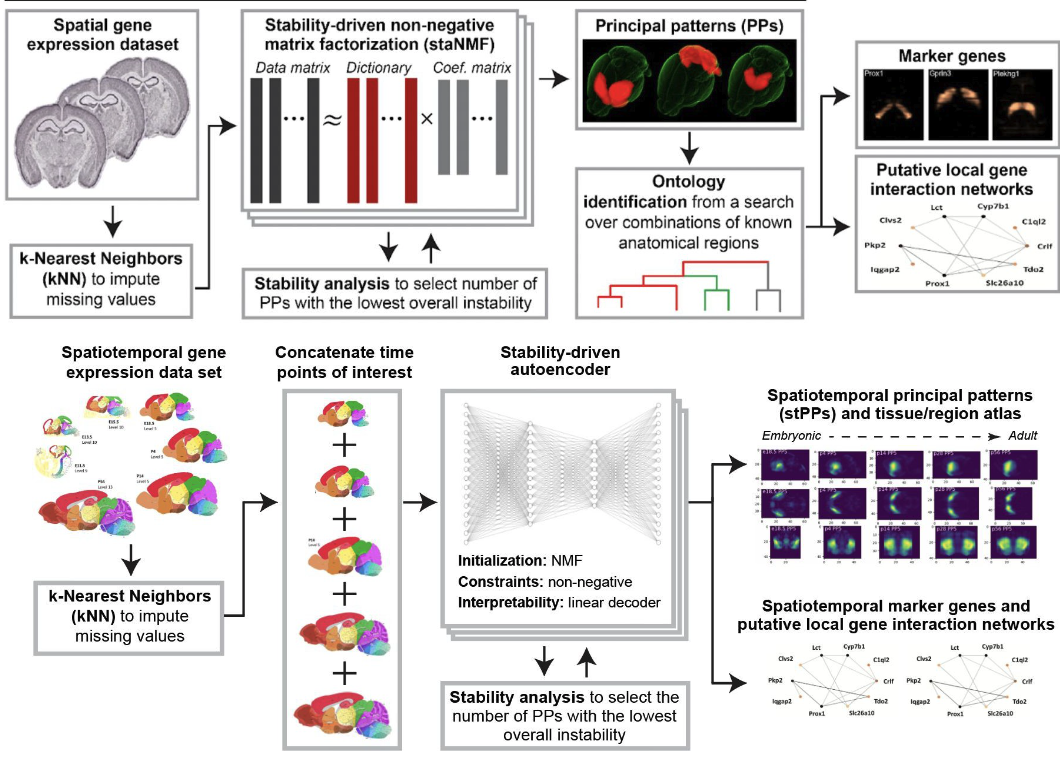
At the Abbasi Lab, I leveraged deep autoencoders, non-negative matrix factorization (NMF), and semi-supervised deep CNNs to uncover significant and previously undiscovered spatial genomic patterns in complex brain images, which included a range of modalities such as fMRI and gene expression data. Additionally, I designed and validated interpretable machine learning frameworks and statistical tools, which revolutionized our understanding of cell types, their spatial arrangement, local gene networks, and revealed new patterns in complex brain images across human and mouse models, significantly advancing our knowledge of brain function, development, aging, and their implications for patient health and clinical decision-making. |
|
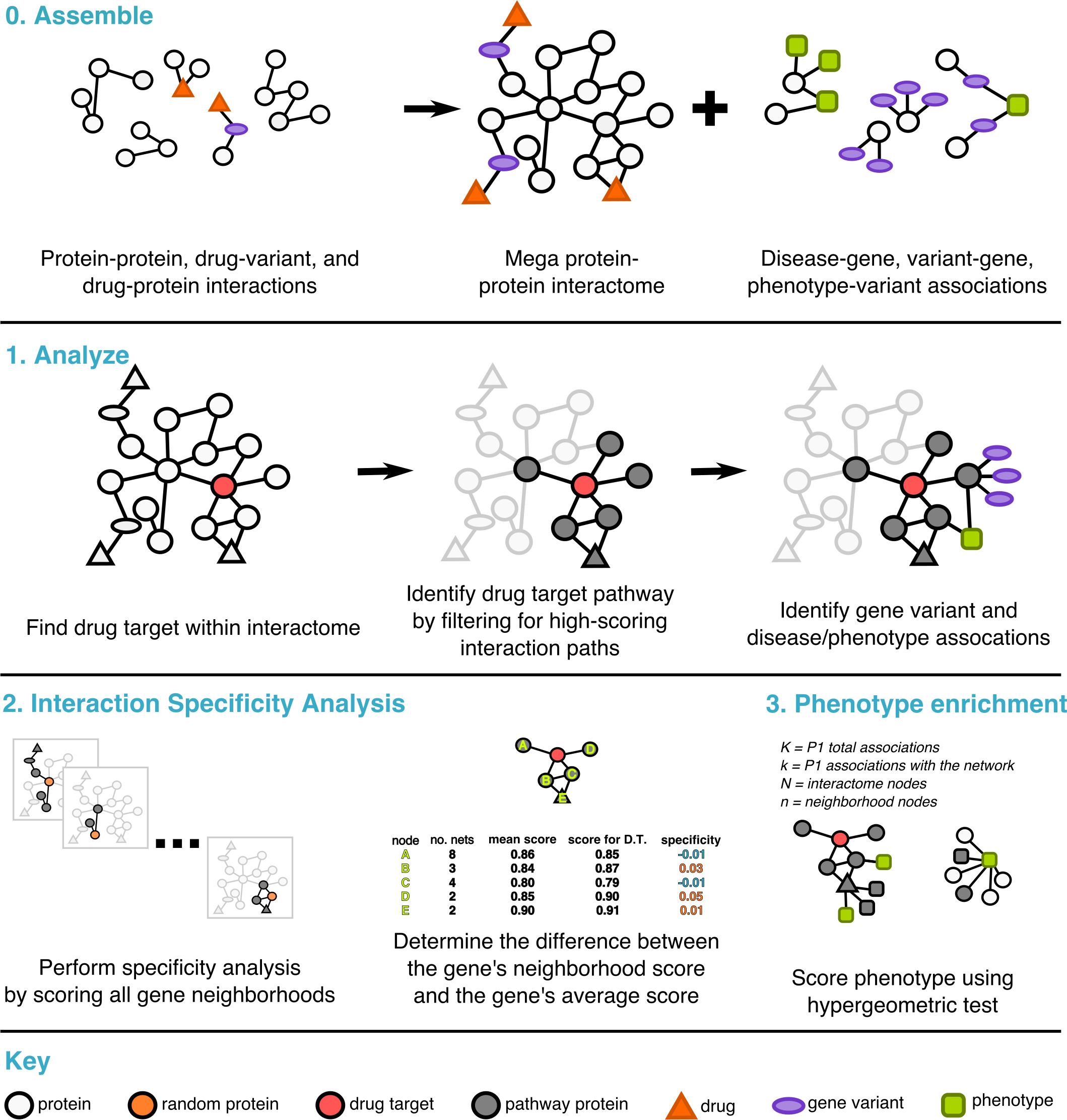
I initiated a research project with Dr. Jenifer Wilson at UCLA using PathFX, a data-driven algorithm that predicts drug adverse effects using protein-protein interaction networks. After training against several tissue and cell type specific models, I developed modifications to the algorithmic pipeline that significantly enhanced PathFX’s capacity to predict drug-induced side effects. My analytical efforts led to the reevaluation of several drug pathways, a crucial step in unveiling new therapeutic strategies and drug repurposing. |
|
During my time at UC Berkeley, I served as a AI and course assistant tutor for several of the EECS and MCB courses. I spent 5 semesters assisting MCB C100A: Biophysical Chemistry - Physical Principles and the Molecules of Life for Professor John Kuriyan (now at Vanderbilt University) and Professor David Savage. |

|
Data C100: Principles and Techniques of Data Science
CS 70: Discrete Mathematics and Probability Theory
|

|
MCB C100A: Biophysical Chemistry - Physical Principles and the Molecules of Life
|
|
|